What it is
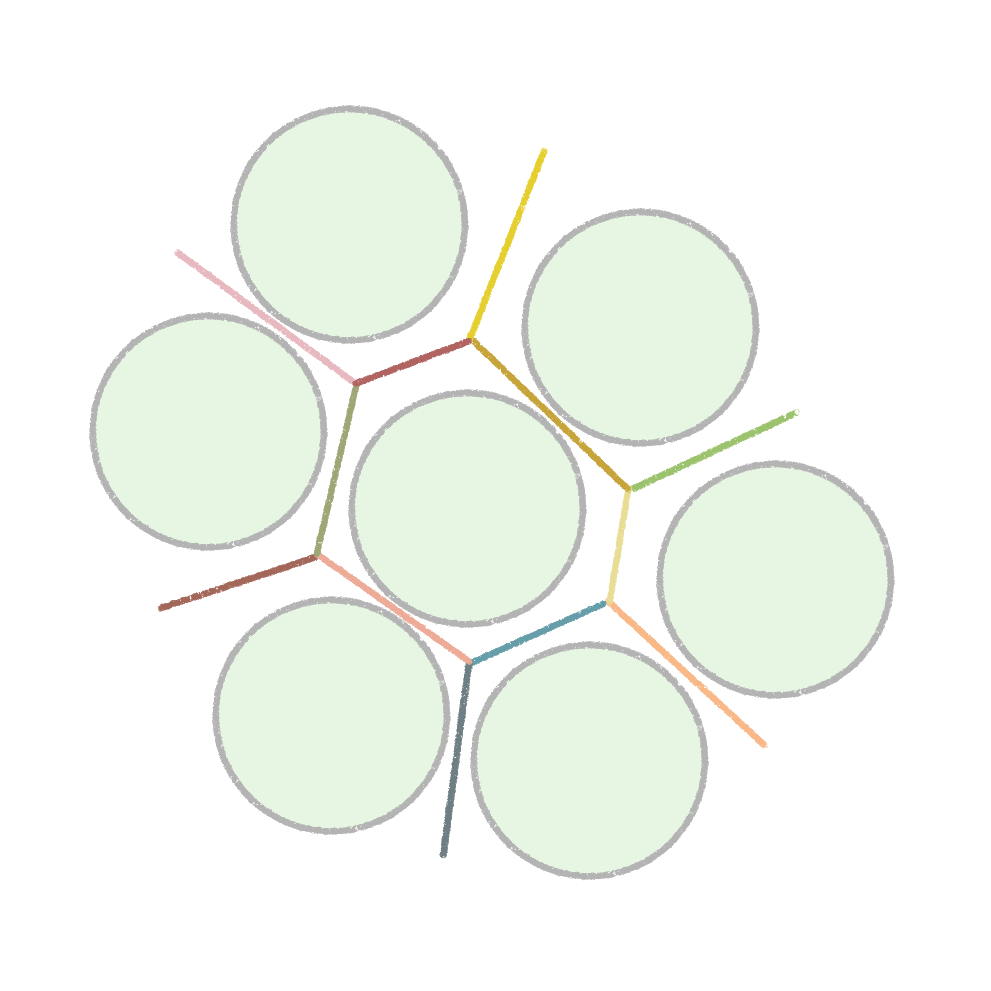
LOVAMAP stands for LOcal Void Analysis using Medial Axis by Particle configuration, and it’s a custom piece of software we’ve developed to analyze important geometrical features of our MAP material – or any granular material for that matter!
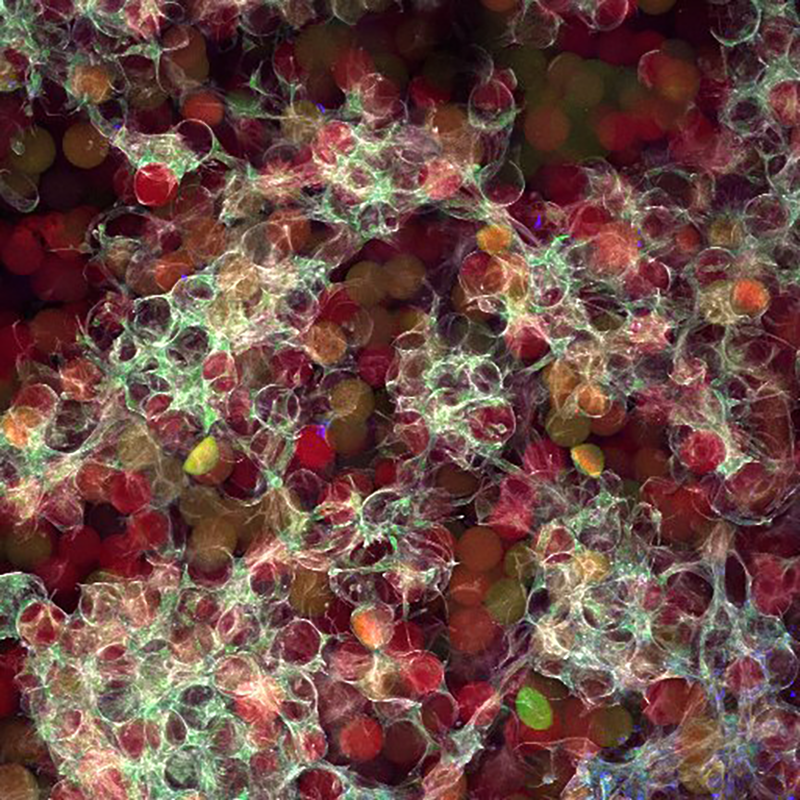
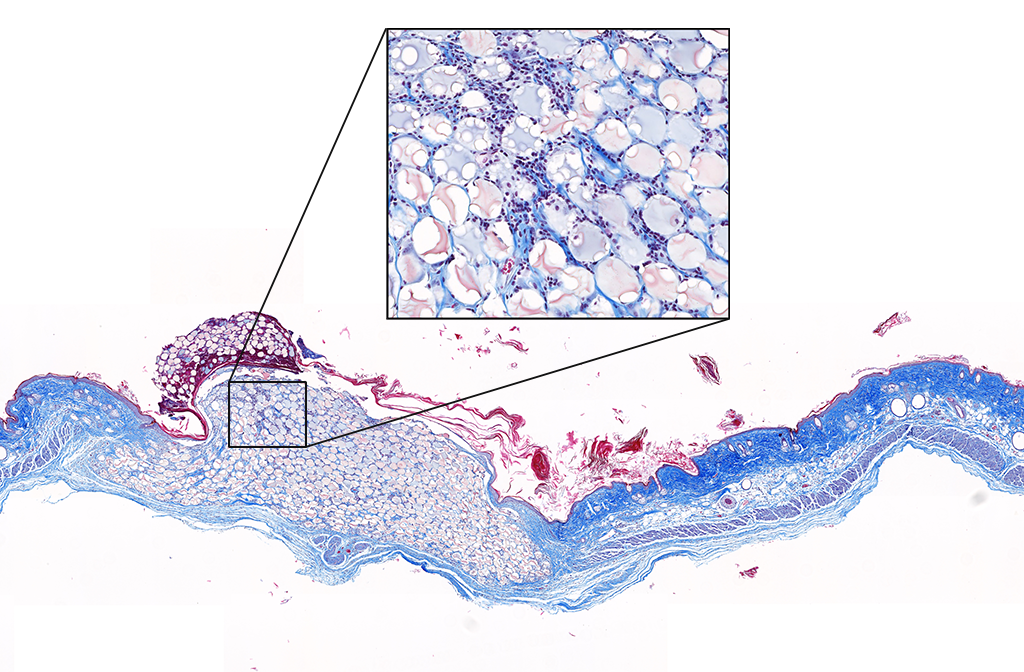
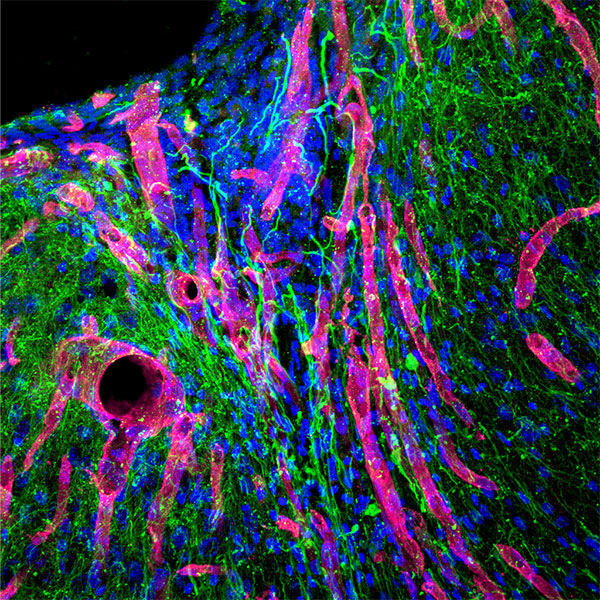
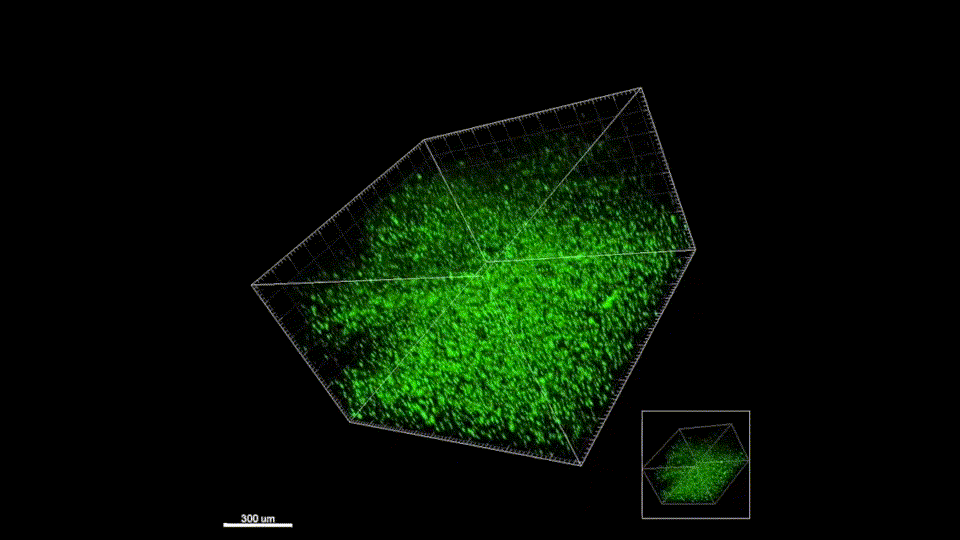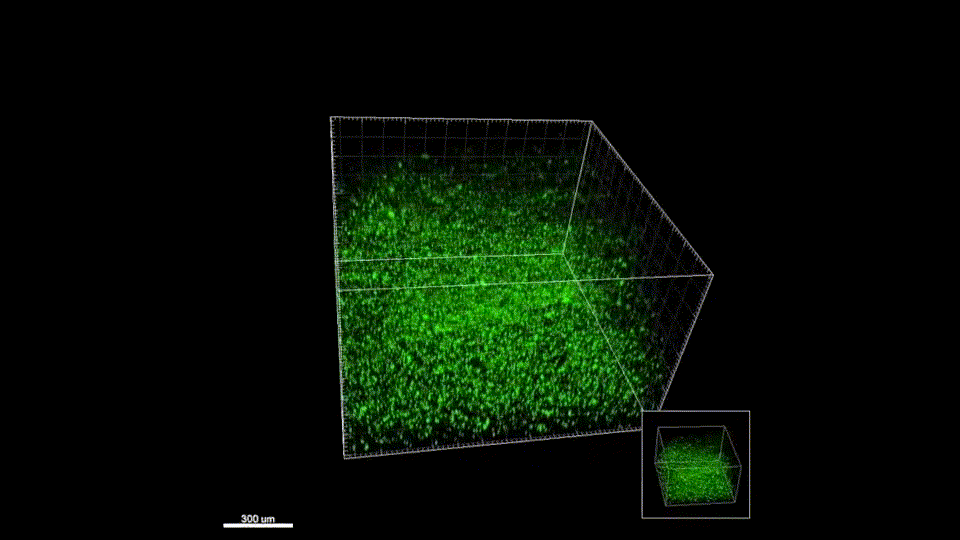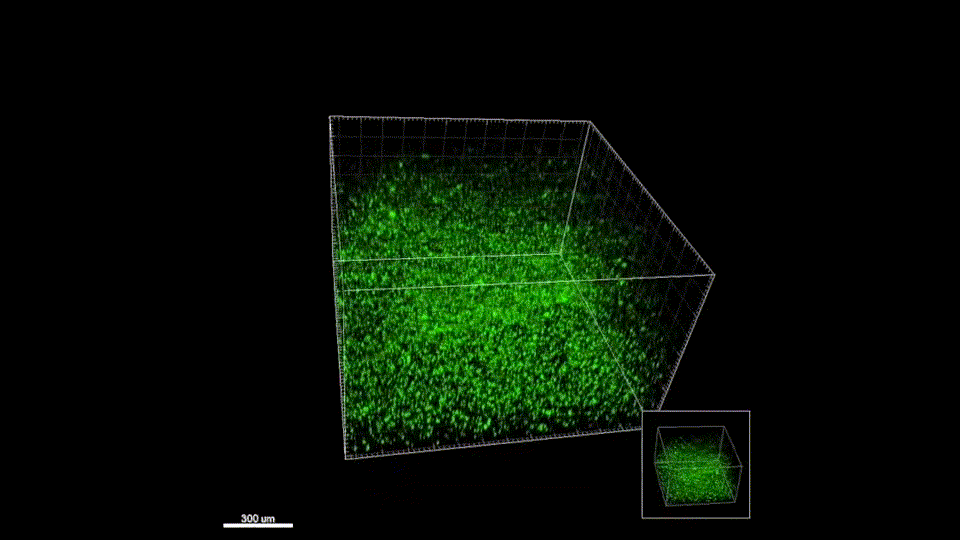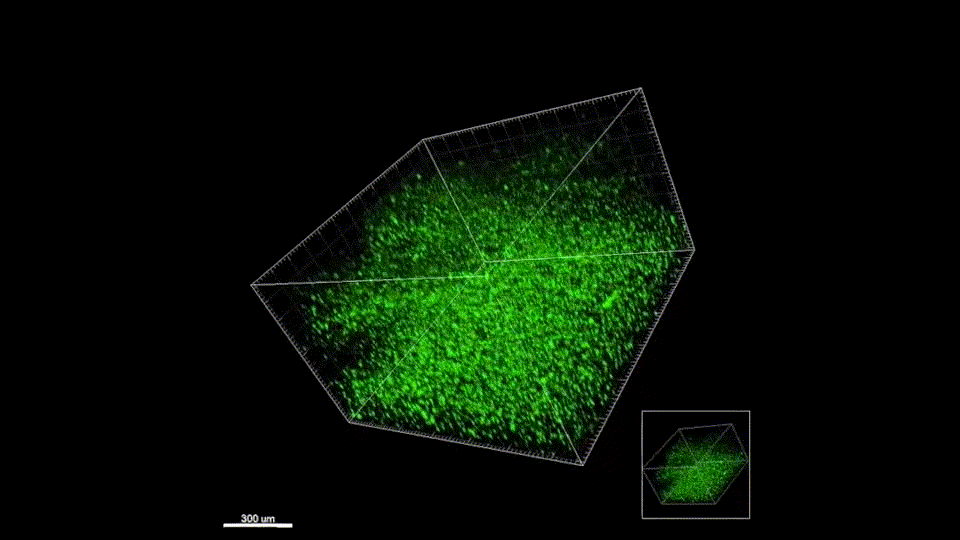
We quickly learned that cells like to hang out inside of MAP scaffolds while they re-build tissue, and that got us thinking – if cells are spending time in the void space between the microparticles, then we should study this space to figure out the best design for promoting tissue repair. But studying the void space among packed particles is a complicated business, and we couldn’t help ourselves – we developed LOVAMAP to go above and beyond when it comes to MAP characterization.
Who helps us
We’ve partnered with Ninjabyte Computing to generate simulated MAP scaffolds that are so realistic, we did a double-take.
You know what’s easier than spending hours in lab fabricating hydrogel microparticles? Pressing a button to simulate hydrogel microparticles. Ninjabyte Computing is equipped with state-of-the-art technology to bring our wildest MAP scaffold fantasies to life. By running LOVAMAP on simulated MAP scaffolds, we can learn more faster. This is why Ninjabyte Computing deserves its very own page here.
How it works
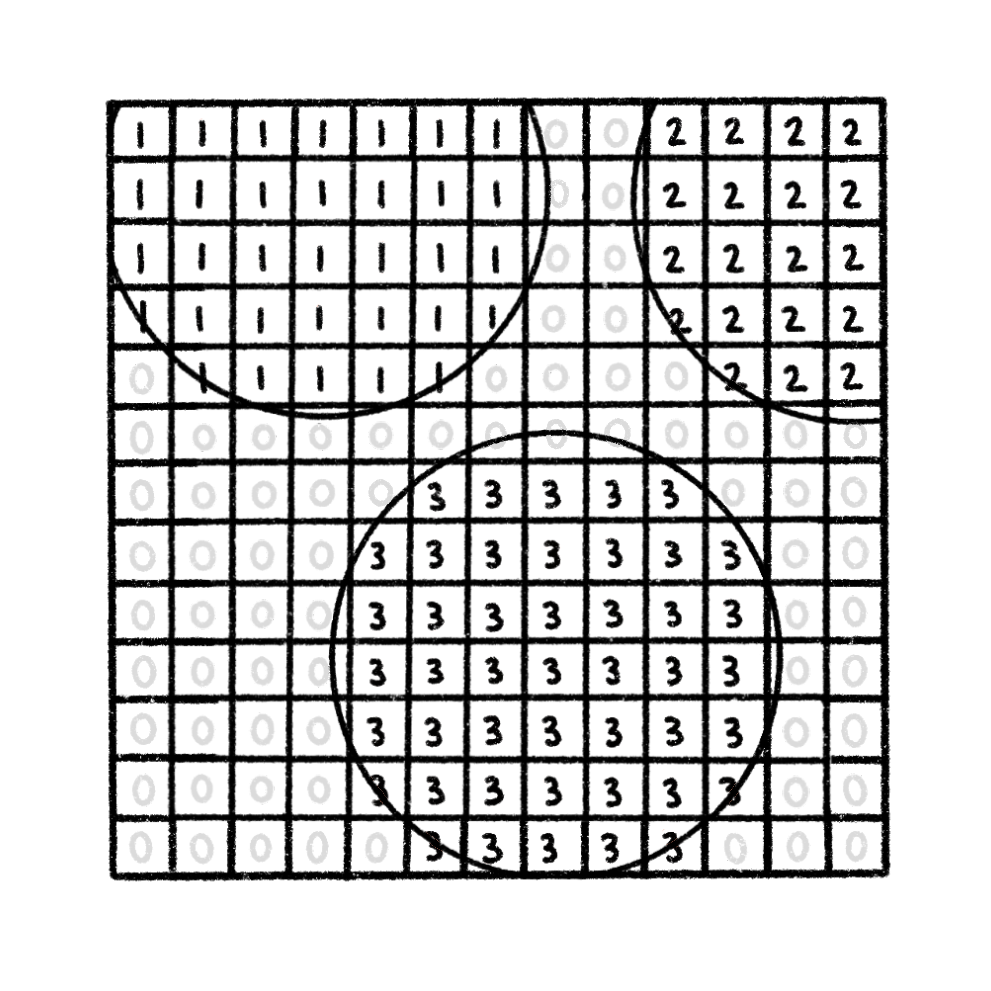
Read in formatted data

Extract spatial landmarks
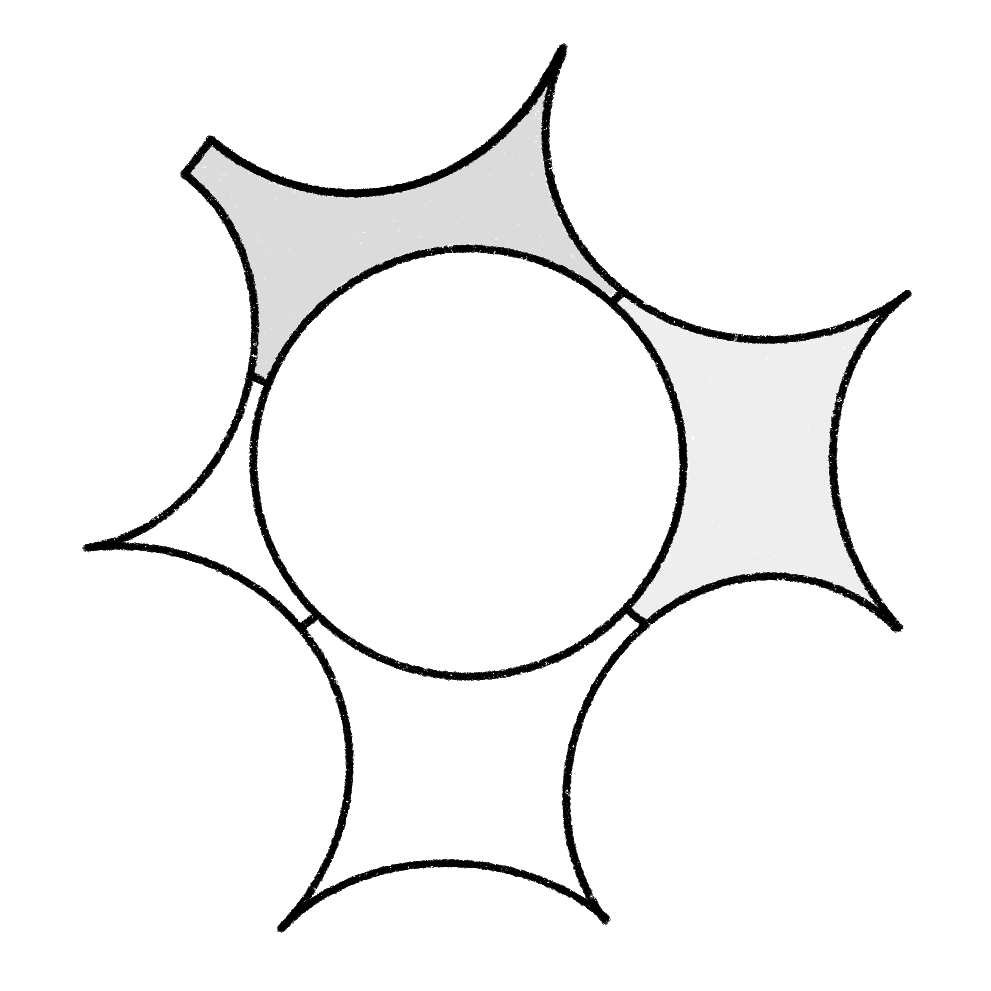
Segment void space
Return descriptor data

Generate images & GIFs
What it offers
LOVAMAP outputs 3 types of descriptors that characterize MAP scaffolds:

GLOBAL DESCRIPTORS

3D PORE DESCRIPTORS